nf-core/variantcall
nf-core/variantcall
NF-Core VariantCall

Using the nf-core template
Indeed, the core principle of nf-core is the standardization of pipeline code structure. By adhering to a generalized pipeline template, all pipelines maintain a consistent design. If you’re looking to build a new nf-core pipeline, the best starting point is this template, which can be accessed via the nf-core create command.
The nf-core create command sets up a new Nextflow pipeline in nf-core style, pre-loaded with nf-core templates and configurations. This provides a strong, standardized foundation upon which you can build your specific pipeline.
Here’s how you can use the command:
nf-core create
- After running this command, You will prompted to enter
Workflow Name,Description,Author. - You will also be asked
if you want to customize which parts of the template are used. Enter ‘No’ for now (but, feel free to explore which parts of the template can be customized). This will Initialise a pipeline git repository in your current directory. Once you’ve provided these details, the command will generate a new pipeline directory with all the necessary files and structure in place, following nf-core guidelines.
,--./,-. ___ __ __ __ ___ /,-._.--~\ |\ | |__ __ / ` / \ |__) |__ } { | \| | \__, \__/ | \ |___ \`-._,-`-, `._,._,' nf-core/tools version 2.8 - https://nf-co.re ? Workflow name variantcall ? Description simple variant calling pipeline ? Author sateesh peri ? Do you want to customize which parts of the template are used? No INFO Creating new nf-core pipeline: 'nf-core/variantcall' INFO Initialising pipeline git repository INFO Done. Remember to add a remote and push to GitHub: cd /home/speri/github/test_slack/nf-core-variantcall git remote add origin git@github.com:USERNAME/REPO_NAME.git git push --all origin INFO This will also push your newly created dev branch and the TEMPLATE branch for syncing. INFO !!!!!! IMPORTANT !!!!!! If you are interested in adding your pipeline to the nf-core community, PLEASE COME AND TALK TO US IN THE NF-CORE SLACK BEFORE WRITING ANY CODE! Please read: https://nf-co.re/developers/>adding_pipelines#join-the-community
cd nf-core-variantcall/
ls
NF-Core Modules
NF-Core Modules are a key feature of the nf-core ecosystem, leveraging the modularity provided by Nextflow’s DSL2 syntax. This allows workflows, subworkflows, and modules to be defined and imported into a pipeline. This modular structure promotes the reuse and sharing of pipeline processes among nf-core pipelines.
The shared modules are stored in the nf-core/modules repository. The modules in this repository are designed to be as atomic as possible, generally encapsulating a single tool. If a tool consists of several subtools (e.g., bwa index and bwa mem), these are stored in individual modules, following the naming convention tool/subtool.
Each module defines the input and output channels, the process script, and the software packaging for a specific process. Conda environments, Docker or Singularity containers are defined within each module. Most of these modules rely on the biocontainers project, which provides single-tool containers for each module.
The nf-core/modules repository also includes tests for each module. These tests run on minimal test data from the nf-core/test-datasets repository (modules branch). These module tests run similarly to pipeline tests on GitHub actions, ensuring that modules are always functional and produce the desired results.
NF-core tools include a series of subcommands for working with nf-core modules.
- To list all the modules available in the
nf-core/modulesrepository, use the following command:
nf-core modules list remote
- To list all the currently installed modules in your local pipeline, use:
nf-core modules list local
This modular approach significantly improves the maintainability and scalability of nf-core pipelines, promoting code reuse and standardization across different pipelines.
The template pipeline comes with
fastqc,multiqcandcustom/dumpsoftwareversionspre-installed
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
INFO Modules installed in '.':
┏━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━┓
┃ Module Name ┃ Repository ┃ Version SHA ┃ Message ┃ Date ┃
┡━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━┩
│ custom/dumpsof… │ https://github.… │ 76cc4938c1f6ea… │ give execution │ 2023-04-28 │
│ │ │ │ permissions to │ │
│ │ │ │ dumpsoftwarever… │ │
│ │ │ │ (#3347) │ │
│ fastqc │ https://github.… │ c8e35eb2055c09… │ Bulk change │ 2022-12-13 │
│ │ │ │ conda syntax for │ │
│ │ │ │ all modules │ │
│ │ │ │ (#2654) │ │
│ multiqc │ https://github.… │ f2d63bd5b68925… │ fix meta.ymls │ 2023-04-28 │
│ │ │ │ for dumpsoftware │ │
│ │ │ │ and multiqc │ │
│ │ │ │ (#3345) │ │
└─────────────────┴──────────────────┴─────────────────┴──────────────────┴────────────┘
- Create a samplesheet from untrimmed-reads:
samplesheet.csvpaste the following and save
sample,fastq_1,fastq_2
SRR2584863,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2584863_1.fastq.gz,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2584863_2.fastq.gz
SRR2584866,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2584866_1.fastq.gz,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2584866_2.fastq.gz
SRR2589044,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2589044_1.fastq.gz,/workspace/nextflow_tutorial/data/untrimmed_fastq/SRR2589044_2.fastq.gz
- Change the
max_memoryandmax_cpusavailable innextflow.configfile:
max_memory = '6.GB'
max_cpus = 4
- To run the workflow with
fastqc,multiqcanddumpsoftwareversionsprocesses
nextflow run main.nf --input samplesheet.csv --fasta /workspace/nextflow_tutorial/data/ref_genome/ecoli_rel606.fasta -profile docker
N E X T F L O W ~ version 21.10.6 Launching `main.nf` [admiring_torvalds] - revision: 55a600f0ab ------------------------------------------------------ ,--./,-. ___ __ __ __ ___ /,-._.--~' |\ | |__ __ / ` / \ |__) |__ } { | \| | \__, \__/ | \ |___ \`-._,-`-, `._,._,' nf-core/variantcall v1.0dev ------------------------------------------------------ Core Nextflow options runName : admiring_torvalds containerEngine: docker launchDir : /workspace/nextflow_tutorial/nf-core-variantcall workDir : /workspace/nextflow_tutorial/nf-core-variantcall/work projectDir : /workspace/nextflow_tutorial/nf-core-variantcall userName : gitpod profile : docker configFiles : /workspace/nextflow_tutorial/nf-core-variantcall/nextflow.config Input/output options input : samplesheet.csv Reference genome options fasta : /workspace/nextflow_tutorial/data/ref_genome/ecoli_rel606.fasta Max job request options max_cpus : 4 max_memory : 12.GB !! Only displaying parameters that differ from the pipeline defaults !! ------------------------------------------------------ If you use nf-core/variantcall for your analysis please cite: * The nf-core framework https://doi.org/10.1038/s41587-020-0439-x * Software dependencies https://github.com/nf-core/variantcall/blob/master/CITATIONS.md ------------------------------------------------------ executor > local (6) [60/43ab20] process > NFCORE_VARIANTCALL:VARIANTCALL:INPUT_CHECK:SAMPLESHEET_CHECK (samplesheet.csv) [100%] 1 of 1 ✔ [27/2aadda] process > NFCORE_VARIANTCALL:VARIANTCALL:FASTQC (SRR2589044_T1) [100%] 3 of 3 ✔ [26/e03e88] process > NFCORE_VARIANTCALL:VARIANTCALL:CUSTOM_DUMPSOFTWAREVERSIONS (1) [100%] 1 of 1 ✔ [de/019672] process > NFCORE_VARIANTCALL:VARIANTCALL:MULTIQC [100%] 1 of 1 ✔ -[nf-core/variantcall] Pipeline completed successfully- WARN: To render the execution DAG in the required format it is required to install Graphviz -- See http://www.graphviz.org for more info. Completed at: 21-Feb-2022 14:46:30 Duration : 1m 55s CPU hours : 0.1 Succeeded : 6
- To clean any previous nextflow runs:
rm -fr results/ work/ .nextflow .nextflow.log*
Adding nf-core modules to a pipeline
To filter the search:
nf-core modules list remote | grep bwa
nf-core modules list remote | grep samtools
nf-core modules list remote | grep bcftools
Adding nf-core modules to a pipeline, if the modules already exist in the nf-core modules repository, can be done with the following command (executing it in the main pipeline directory):
nf-core modules install <module name>
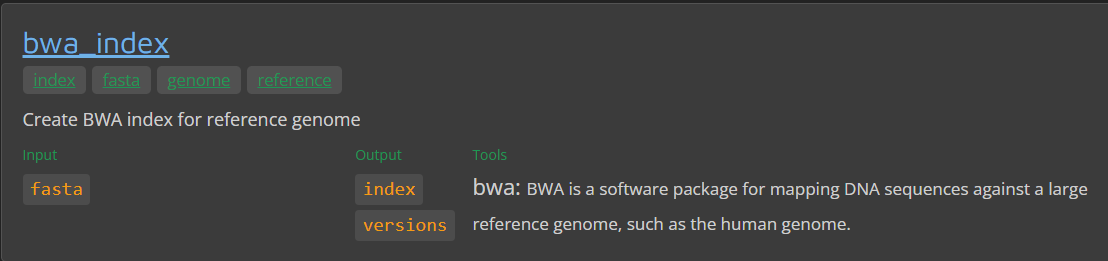
nf-core modules install bwa/index
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
INFO Installing 'bwa/index'
INFO Use the following statement to include this module:
include { BWA_INDEX } from '../modules/nf-core/bwa/index/main'
nf-core modules install bwa/mem
,--./,-.
___ __ __ __ ___ /,-._.--~\
|\ | |__ __ / ` / \ |__) |__ } {
| \| | \__, \__/ | \ |___ \`-._,-`-,
`._,._,'
nf-core/tools version 2.8 - https://nf-co.re
INFO Installing 'bwa/mem'
INFO Use the following statement to include this module:
include { BWA_MEM } from '../modules/nf-core/bwa/mem/main'
nf-core modules install samtools/sort
nf-core modules install samtools/index
nf-core modules install bcftools/mpileup
NF-core modules are stored under the modules/nf-core directory. To use these modules in your main pipeline workflow (e.g., workflows/<pipeline-name>.nf) or a sub-workflow, you need to include them in the corresponding Nextflow file using the include statement:
include { TOOL_SUBTOOL } from '../modules/nf-core/modules/<tool/subtool>/main'
For example, if you’re working with a pipeline named nf-core-variantcall, you could add the following includes in your nf-core-variantcall/workflows/variantcall.nf:
include { BWA_INDEX } from '../modules/nf-core/modules/bwa/index/main'
include { BWA_MEM } from '../modules/nf-core/modules/bwa/mem/main'
include { SAMTOOLS_SORT } from '../modules/nf-core/modules/samtools/sort/main'
include { SAMTOOLS_INDEX } from '../modules/nf-core/modules/samtools/index/main'
include { BCFTOOLS_MPILEUP } from '../modules/nf-core/modules/bcftools/mpileup/main'
Then, within the workflow scope of the same variantcall.nf file, you can call these modules as follows:
//
// MODULE: Run BWA-INDEX
//
BWA_INDEX(params.fasta)
//
// MODULE: Run BWA-ALIGN
//
BWA_MEM(INPUT_CHECK.out.reads, BWA_INDEX.out.index, "view")
//
// MODULE: Run SAMTOOLS-SORT
//
SAMTOOLS_SORT(BWA_MEM.out.bam)
//
// MODULE: Run SAMTOOLS-INDEX
//
SAMTOOLS_INDEX(SAMTOOLS_SORT.out.bam)
Module-specific options, like tool options, can be defined in the nf-core-variantcall/conf/modules.config configuration file. For instance, you might want to add the following configuration to the modules.config file:
withName: SAMTOOLS_SORT {
ext.prefix = "sorted"
}
Once you’ve set up your pipeline with the appropriate modules and configurations, you can run the pipeline using the nextflow run command:
nextflow run main.nf --input samplesheet.csv --fasta /workspace/nextflow_tutorial/data/ref_genome/ecoli_rel606.fasta -profile docker
This command instructs Nextflow to run your pipeline (defined in main.nf), specifying input and reference fasta file locations, and telling it to use Docker containers for software dependencies (with -profile docker).
Output
N E X T F L O W ~ version 21.10.6 Launching `main.nf` [magical_einstein] - revision: 55a600f0ab ------------------------------------------------------ ,--./,-. ___ __ __ __ ___ /,-._.--~' |\ | |__ __ / ` / \ |__) |__ } { | \| | \__, \__/ | \ |___ \`-._,-`-, `._,._,' nf-core/variantcall v1.0dev ------------------------------------------------------ Core Nextflow options runName : magical_einstein containerEngine: docker launchDir : /workspace/nextflow_tutorial/nf-core-variantcall workDir : /workspace/nextflow_tutorial/nf-core-variantcall/work projectDir : /workspace/nextflow_tutorial/nf-core-variantcall userName : gitpod profile : docker configFiles : /workspace/nextflow_tutorial/nf-core-variantcall/nextflow.config Input/output options input : samplesheet.csv Reference genome options fasta : /workspace/nextflow_tutorial/data/ref_genome/ecoli_rel606.fasta Max job request options max_cpus : 4 max_memory : 6.GB !! Only displaying parameters that differ from the pipeline defaults !! ------------------------------------------------------ If you use nf-core/variantcall for your analysis please cite: * The nf-core framework https://doi.org/10.1038/s41587-020-0439-x * Software dependencies https://github.com/nf-core/variantcall/blob/master/CITATIONS.md ------------------------------------------------------ executor > local (16) [3f/2fccb5] process > NFCORE_VARIANTCALL:VARIANTCALL:INPUT_CHECK:SAMPLESHEET_CHECK (samplesheet.csv) [100%] 1 of 1 ✔ [23/776fe6] process > NFCORE_VARIANTCALL:VARIANTCALL:FASTQC (SRR2584863_T1) [100%] 3 of 3 ✔ [2e/2d108b] process > NFCORE_VARIANTCALL:VARIANTCALL:CUSTOM_DUMPSOFTWAREVERSIONS (1) [100%] 1 of 1 ✔ [59/7e75bb] process > NFCORE_VARIANTCALL:VARIANTCALL:BWA_INDEX (ecoli_rel606.fasta) [100%] 1 of 1 ✔ [ce/50d7ee] process > NFCORE_VARIANTCALL:VARIANTCALL:BWA_MEM (SRR2584866_T1) [100%] 3 of 3 ✔ [56/dae396] process > NFCORE_VARIANTCALL:VARIANTCALL:SAMTOOLS_SORT (SRR2584866_T1) [100%] 3 of 3 ✔ [90/5ac8a9] process > NFCORE_VARIANTCALL:VARIANTCALL:SAMTOOLS_INDEX (SRR2584866_T1) [100%] 3 of 3 ✔ [40/2df600] process > NFCORE_VARIANTCALL:VARIANTCALL:MULTIQC [100%] 1 of 1 ✔ -[nf-core/variantcall] Pipeline completed successfully- WARN: To render the execution DAG in the required format it is required to install Graphviz -- See http://www.graphviz.org for more info. Completed at: 04-Mar-2022 01:57:43 Duration : 5m 30s CPU hours : 0.3 Succeeded : 16