Introduction to NextFlow
Nextflow Introduction
Learning Objectives:
- Understand what a workflow management system is.
- Understand the benefits of using a workflow management system.
- Explain the benefits of using Nextflow as part of your bioinformatics workflow.
- Explain the components of a Nextflow script.
- Run a Nextflow script.
Workflows
-
Data analysis involves a series of tasks, such as gathering, cleaning, and processing data. These sequences of tasks are referred to as workflows or pipelines.
-
Workflows often require executing multiple software packages, which may run on various computing environments, such as desktops or compute clusters.
-
Traditionally, these workflows have been linked together using scripts in general-purpose programming languages, like Bash or Python.
However, managing programming logic and software becomes challenging as workflows grow larger and more complex.
Workflow Management Systems
Recently, Workflow Management Systems (WfMS), such as Snakemake, Galaxy, and Nextflow, have emerged to specifically manage computational data-analysis workflows.
These Workflow Management Systems offer multiple features that simplify the development, monitoring, execution, and sharing of pipelines:
- Run time management
- Software management
- Portability & Interoperability
- Reproducibility
- Re-entrancy
Wratten, L., Wilm, A. & Göke, J. Reproducible, scalable, and shareable analysis pipelines with bioinformatics workflow managers. Nat Methods 18, 1161–1168 (2021). https://doi.org/10.1038/s41592-021-01254-9
Core Features of Nextflow:
-
Fast Prototyping: Nextflow’s simple syntax allows for the reuse of existing scripts and tools, enabling rapid pipeline development.
-
Reproducibility: With support for various container technologies like Docker and Singularity, as well as the package manager Conda, Nextflow facilitates the creation of self-contained pipelines. Integration with GitHub/GitLab enables efficient version management and the reproduction of any previous configuration.
-
Portability: Nextflow’s syntax separates functional logic (workflow steps) from execution settings (how the workflow runs), allowing the pipeline to be executed on multiple platforms (e.g., local compute, HPC clusters, or cloud services like AWS) without altering workflow steps.
-
Simple Parallelism: Built on the dataflow programming model, Nextflow greatly simplifies parallel task execution.
-
Continuous Checkpoints: Nextflow automatically tracks all intermediate results produced during pipeline execution. This feature allows users to resume execution from the last successfully executed step, regardless of the cause of interruption.
Nextflow Basic Concepts
Processes, Channels, and Workflows
Nextflow workflows consist of three primary components: processes, channels, and workflows.
-
Processesdefine tasks to be executed. Process scripts can be written in any scripting language compatible with the Linux platform (Bash, Perl, Ruby, Python, etc.). Processes create a task for each complete input set, with each task executing independently and without interaction with other tasks. Data can only be passed between process tasks via asynchronous queues calledChannels. -
Processes specify
inputsandoutputsfor a task. Channels manipulate the flow of data between processes. The interaction between processes, and ultimately the pipeline execution flow, is explicitly defined in aworkflowsection. -
In the example below, a channel contains three elements, such as three data files. A process receives the channel as input. Since the channel contains three elements, three independent instances (tasks) of the process run in parallel. Each task generates an output, which is passed to another channel and serves as input for the next process.
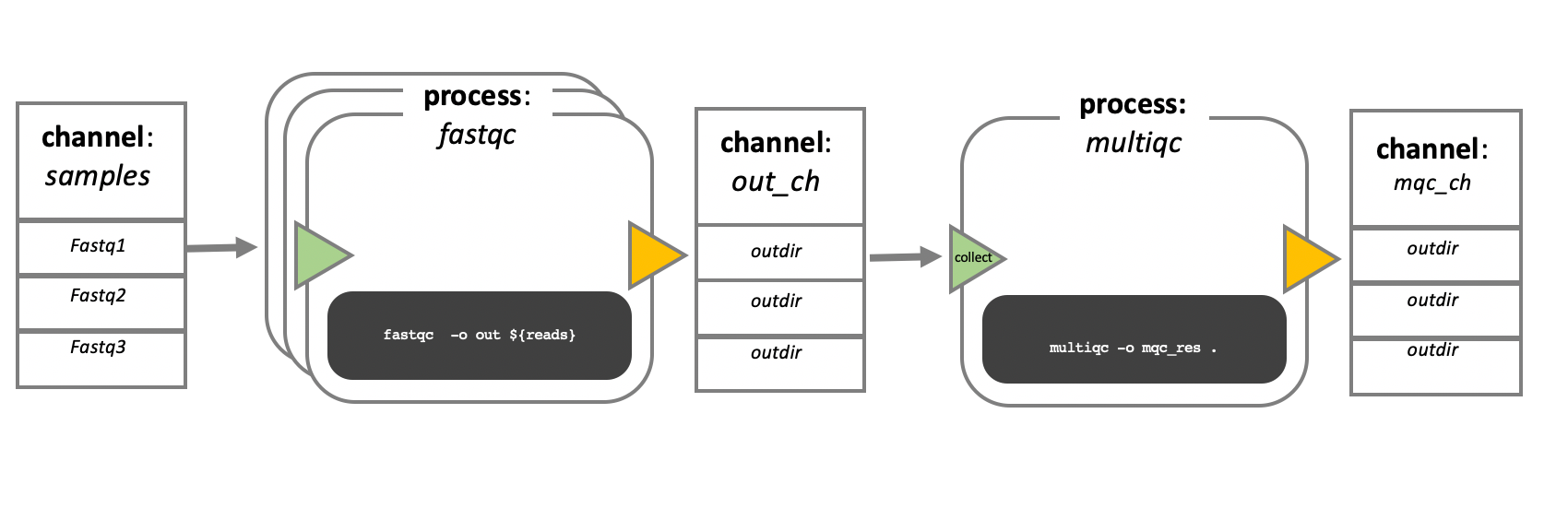
Workflow Execution
-
While a process determines the command or script to be executed, the executor defines how that script is run on the target system.
-
By default, processes execute on the local computer. The local executor is useful for pipeline development, testing, and small-scale workflows. However, large-scale computational pipelines often require High Performance Clusters (HPC) or Cloud platforms.
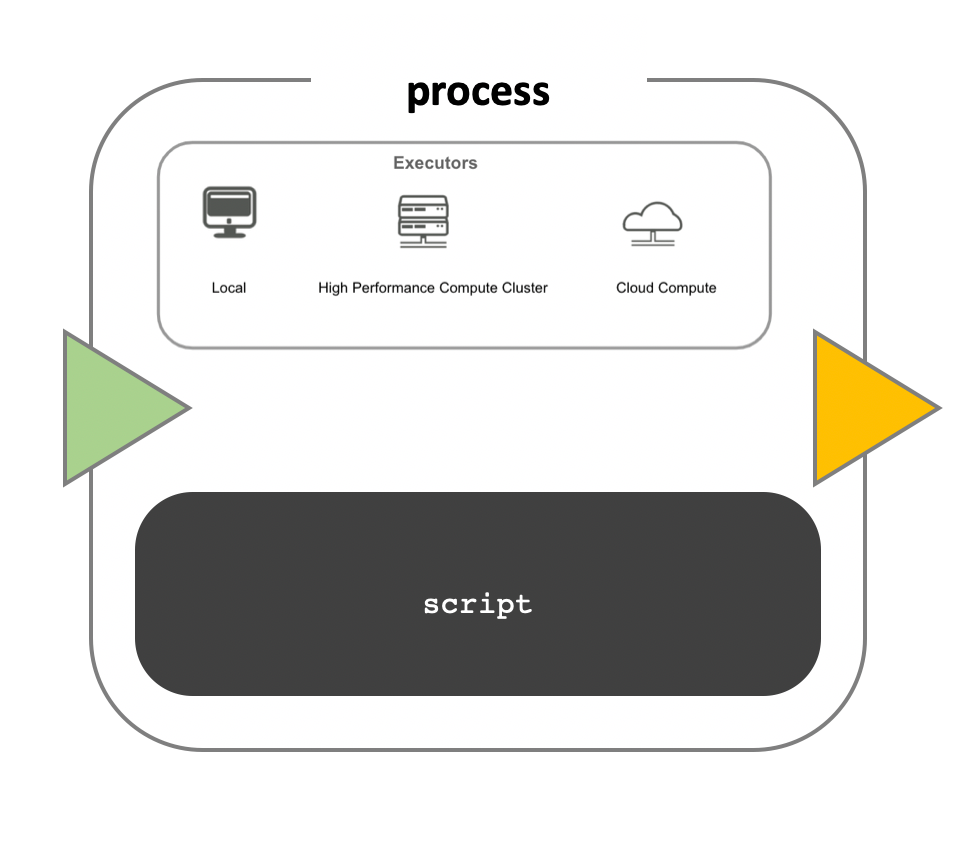
-
Nextflow separates the pipeline’s functional logic from the underlying execution platform. This allows a pipeline to be written once and executed on a local computer, compute cluster, or the cloud without modifying the workflow. The target execution platform can be defined in a configuration file.
-
Nextflow offers out-of-the-box support for major batch schedulers and cloud platforms, such as Sun Grid Engine, SLURM job scheduler, AWS Batch service, and Kubernetes. A comprehensive list can be found here.
Your First Nextflow Script
Navigate to the Nextflow tutorial directory:
cd /workspace/nextflow_tutorial
Now, create a Nextflow script that counts the number of lines in a file. To do this, create the file word_count.nf in the current directory using code word_count.nf or your preferred text editor and copy-paste the following code:
#!/usr/bin/env nextflow
nextflow.enable.dsl=2
/*
========================================================================================
Workflow parameters are written as params.<parameter>
and can be initialised using the `=` operator.
========================================================================================
*/
params.input = "data/untrimmed_fastq/SRR2584863_1.fastq.gz"
/*
========================================================================================
Input data is received through channels
========================================================================================
*/
input_ch = Channel.fromPath(params.input)
/*
========================================================================================
Main Workflow
========================================================================================
*/
workflow {
// The script to execute is called by its process name, and input is provided between brackets.
NUM_LINES(input_ch)
/* Process output is accessed using the `out` channel.
The channel operator view() is used to print process output to the terminal. */
NUM_LINES.out.view()
}
/*
========================================================================================
A Nextflow process block. Process names are written, by convention, in uppercase.
This convention is used to enhance workflow readability.
========================================================================================
*/
process NUM_LINES {
input:
path read
output:
stdout
script:
"""
# Print reads
printf '${read}\t'
# Unzip file and count number of lines
gunzip -c ${read} | wc -l
"""
}
This Nextflow script includes:
- An optional interpreter directive (“
Shebang”) line, specifying the location of the Nextflow interpreter. nextflow.enable.dsl=2to enable DSL2 syntax.- A multi-line Nextflow comment, written using C-style block comments, followed by a single-line comment.
- A pipeline parameter
params.inputwith a default value, which is the relative path to the location of a compressed fastq file, as a string. - A Nextflow process block named
NUM_LINES, which defines what the process does. - An
inputdefinition block that assigns theinputto the variableread, and declares that it should be interpreted as a file path. - An
outputdefinition block that uses the Linux/Unix standard output streamstdoutfrom the script block. - A script block containing the bash commands
printf '${read}'andgunzip -c ${read} | wc -l. - A Nextflow channel
input_chused to read in data for the workflow. - An unnamed
workflowexecution block, which is the default workflow to run. - A call to the process
NUM_LINESwith the input channelinput_ch. - An operation on the process output, using the channel operator
.view().
Run a Nextflow Script
To run the script, enter the following command in your terminal:
nextflow run word_count.nf
The output should resemble the text shown below:
N E X T F L O W ~ version 21.04.3
Launching `word_count.nf` [marvelous_mestorf] - revision: c09ee14ad4
executor > local (1)
[81/92e3e9] process > NUM_LINES (1) [100%] 1 of 1 ✔
SRR2584863_1.fastq.gz 6213036
- The first line shows the Nextflow version number.
- The second line displays the run name marvelous_mestorf (adjective and scientist name) and revision id c09ee14ad4.
- The third line indicates that the process has been executed locally (executor > local).
- The following line shows the process id
81/92e3e9, process name, percentage task completion, and the number of instances of the process that have been run. - The final line is the standard output of the process displayed using the
.viewoperator.
Pipeline Parameters
-
In the Nextflow
word_count.nfscript, a pipeline parameterparams.inputis defined. Pipeline parameters allow you to modify the workflow input at runtime, through the command line or a configuration file, so it’s not hard-coded into the script. -
Pipeline parameters are declared in the workflow by appending the
paramsprefix, separated by a dot character, to a variable name, e.g.,params.input. You can specify their value on the command line by prefixing the parameter name with a double dash character, e.g.,--input. -
Wildcards can also be used to specify multiple input files. In the example below, the
*is used to match any sequence of characters with.fastq.gz. Note: When using wildcard characters on the command line, you must enclose the value in quotes.
Re-run the Nextflow script by entering the following command in your terminal:
nextflow run word_count.nf --input 'data/untrimmed_fastq/*.fastq.gz'
The string specified on the command line will override the default value of the parameter in the script. The output will look like this:
N E X T F L O W ~ version 21.04.3 Launching `word_count.nf` [desperate_agnesi] - revision: bf77afb9d7 executor > local (6) [60/584db8] process > NUM_LINES (5) [100%] 6 of 6 ✔ SRR2589044_1.fastq.gz 4428360 SRR2589044_2.fastq.gz 4428360 SRR2584863_1.fastq.gz 6213036 SRR2584863_2.fastq.gz 6213036 SRR2584866_2.fastq.gz 11073592 SRR2584866_1.fastq.gz 11073592
The pipeline executes the process 6 times; one process for each file matching the string. Since each process is executed in parallel, there is no guarantee of which output is reported first. When you run this script, you may see the process output in a different order.
- Nextflow stores intermediate files in a
worksub-directory in your current directory.
work/
├── 13
│ └── 46936e3927b74ea6e5555ce7b7b56a
│ └── SRR2589044_2.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2589044_2.fastq.gz
├── 43
│ └── 3819a4bc046dd7dc528de8eae1c6b8
│ └── SRR2584863_1.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2584863_1.fastq.gz
├── 5a
│ └── d446a792db2781ccd0c7aaafdff329
│ └── SRR2584866_2.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2584866_2.fastq.gz
├── 76
│ └── b1e11f2e706b901e0c57768a2af59f
│ └── SRR2589044_1.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2589044_1.fastq.gz
├── 9c
│ └── 1b1ebc2ea11a395e4e9dcf805b2c7d
│ └── SRR2584863_2.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2584863_2.fastq.gz
└── ce
└── b94ef609ee54f4b7ea79dc23eb32bb
└── SRR2584866_1.fastq.gz -> nextflow_tutorial/data/untrimmed_fastq/SRR2584866_1.fastq.gz
12 directories, 6 files
- Nextflow has a
logcommand to display the logs of all previous executions.
Type the following on the command line to display an output similar as below
nextflow log
TIMESTAMP DURATION RUN NAME STATUS REVISION ID SESSION ID COMMAND
2021-11-16 07:17:23 5.9s irreverent_leakey OK bf77afb9d7 17d06cd0-3bb9-4d32-9d75-48bfdf5401a9 nextflow run word_count.nf
2021-11-16 07:23:00 11.1s desperate_agnesi OK bf77afb9d7 41f78242-27d7-462a-a88d-80b6ec9dc5db nextflow run word_count.nf --input 'data/untrimmed_fastq/*.fastq.gz'
Quick Recap
- A workflow is a sequence of tasks that process a set of data, and a workflow management system (WfMS) is a computational platform that provides an infrastructure for the set-up, execution and monitoring of workflows.
- Nextflow scripts comprise of
channelsfor controlling inputs and outputs, andprocessesfor defining workflow tasks.- You run a Nextflow script using the
nextflow runcommand.- Nextflow stores working files in the
workdirectory.- The
nextflow logcommand can be used to see information about executed pipelines.